Pioneering neuroscientist Santiago Ramón y Cajal jump-started the search for a “components catalogue” of the human brain towards the end of the 19th century. His intricate drawings of brain cells, complete with their weblike connections, still appear in many textbooks. Looking for brain parts is driven by more than curiosity. Before the generations-long endeavor of deciphering the brain can proceed, neuroscientists need to first identify its multitude of component parts and then figure out what each one does.
The task is complicated by the many ways cells can differ. Cajal provided glimpses of the shapes that distinguish some cell types, but also left a virtually infinite amount of work for future generations of neuroanatomists. Cells can differ by location, biochemistry and other properties. These different descriptors often do not correspond to each other in any simple way, a fact that has fueled debates about how to define cell types. As tools to record the signals neurons use to communicate became available, researchers have tried to categorize cells by comparing their different firing patterns, the specialty of the discipline known as electrophysiology. This effort comes closer to classifying what cells do, but is still descriptive in that it describes behavior rather than morphology.
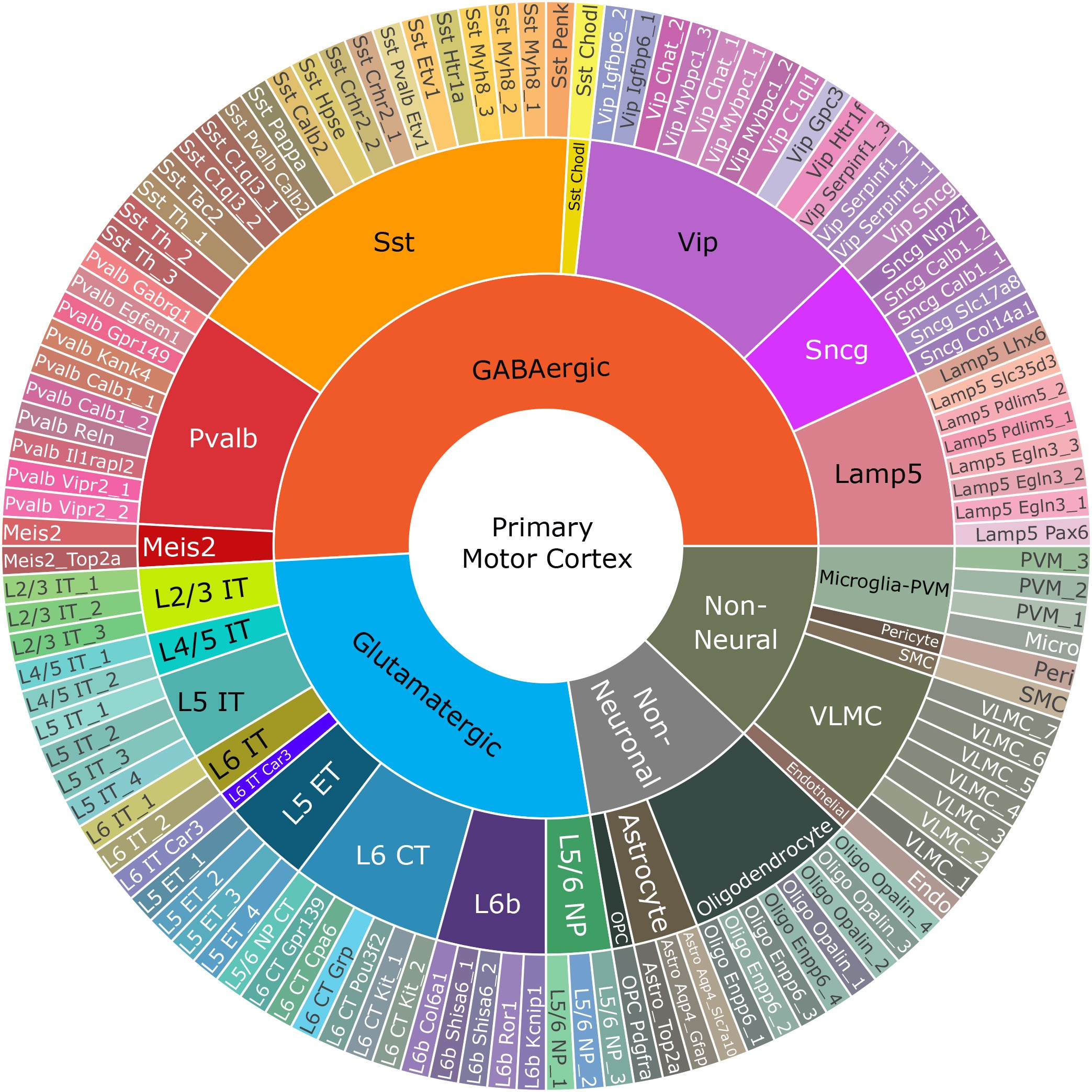
The journey towards a definition that describes cells according to their function comes to an end at the genome, the blueprint that underlies all other biological properties. That these efforts are now bearing fruit is demonstrated by a large, international consortium, funded by the National Institute of Health’s (NIH) BRAIN Initiative. It has produced a genomics-based census of the cell types in one region, the primary motor cortex, responsible for controlling complex movement.
This atlas applies equally to mice, monkeys and humans. The motor cortex became the region of focus as a first step toward more comprehensive brain inventories because it is both well-studied and similar across species. Called the Brain Initiative Cell Census Network (BICCN), the group comprises the efforts of many labs, spearheaded by the Allen Institute for Brain Science, in Seattle. Their findings, described in 17 papers taking over this week’s Nature, represent a resource that will accelerate efforts to understand brain functio, and provide insight into brain diseases and disorders.
The project used the widest range of tools for probing brain cells ever brought to bear in a single, coordinated effort. Studies document how these tools measure different cellular properties, while a flagship paper describes the integration of data from 11 companion papers, to produce a cross-species atlas of cell-types. A few studies push beyond the motor cortex in the mouse to detail other regions and brain networks. Still other studies ask questions about how human brains are shaped, by evolution and during early development.
The research relied heavily on “genomic” technologies, such as “transcriptomics,” which measures gene activity by sequencing RNA molecules in different cell types. Researchers also employed “epigenomic” techniques that look at how gene activity is influenced without altering the underlying genetic code. The researchers used two such techniques that observe how genes are switched on and off by the addition of a chemical group to DNA, or how genes can be read more easily by rearranging the structure DNA is wrapped up in.
The researchers used genomic data to produce a “ground truth” set of classifications for different cell types. They also measured other properties, like shape, and electrophysiology, to add extra dimensions to the genetic categories and begin inspecting how well they align. “There’s a link between genes and properties, so it’s more than just a means to classify, it’s the explanatory basis for what cells do,” says neuroscientist Ed Lein, of the Allen Institute, who helped coordinate the project and led two of the studies. Some studies also used new or recently developed techniques that measure multiple properties simultaneously. “Patch-seq” recorded the electrophysiology and gene activity of individual cells where they are situated before reconstructing their 3-D shape. “Spatial transcriptomics” tools that measure gene activity of cells by combining genomics and brain-imaging allowed the mapping of cells’ locations, providing information about the distribution and proportions of cell types.
Methods for tracing neural connections also enabled the generation of an input/output wiring diagram of the mouse motor cortex. “This concerted effort allowed us to look at the cell types from all different angles,” says neuroscientist Aparna Bhaduri, of University of California, Los Angeles, who led one of the human brain development studies. “Being part of this package means many of these new techniques will have wider applicability, sooner, because they’re so rigorously tested against all the others.”
The data sets, curated by a part of the consortium called the BRAIN Cell Data Center (BCDC), are publicly available. “This is helping to standardize the field. It’s going to be a foundational cell-type classification reference, much like the human genome for genetics,” Lein says. He hopes this will allow researchers to move past a very basic task in brain science, the debating of definitions. “Understanding the components lets the field move to the next set of questions,” he says. “Like what do these cells do?”
The extensive catalogue would not have been possible without a series of technological developments that allows individual brain cells to be poked and probed. “Single-cell genomics is transforming this field, and many other fields of biology,” Lein says. “It has provided a common language for describing cellular diversity.” Bulk tissue analysis has been possible for over a decade, but techniques capable of examining individual cells have only become standardized over the past five years. Measuring gene activity, and regulation, is important, because all cells contain the same DNA, but different cell types implement it differently. “There’s maybe a hundred different cell types in a small patch of your cortex, and we need to understand how each type deploys its genome differently;” says neuroscientist Fenna Krienen, of Harvard Medical School, who worked on the cross-species study. “That’s what single-cell resolution enables, and that enables us to do all sorts of things we couldn’t imagine doing five years ago.”
Combined analyses during the project produced a taxonomy tree, much like “tree of life” illustrations. Major branches reflect important groupings, with shared developmental origins. A first branch separates neural and nonneural cells, splitting off, say, blood cells. The second division, between neuronal and nonneuronal types, separates neurons from “support” cell types, collectively termed “glial cells.” Neurons then split into excitatory types, which increase the chances of other cells firing, and inhibitory types, which put brakes on the activity of other cells. These two broad categories divide into 24 major “subclasses” (including nonneural and glial cell types), which are mostly conserved between species. These can be further divided to arrive at the final branches—the “leaves” of the tree, designated as “t-types,” the “t” being a shortening of “transcriptional,” the genomic means of classifying cell types. The number of these categories differ between species (116 in mice, 127 in humans, 94 in marmosets). The researchers then integrate transcriptomic data from all three species to find 45 t-types that are common, including 24 excitatory, 13 inhibitory and eight nonneuronal cell types, such as astrocytes and oligodendrocytes.
Similarity between species suggests these cell types play important roles in brain function. “Evolutionary conservation is pretty strong evidence of things being under tight genetic control,” Lein says. “And that those elements must therefore be important for the function of the nervous system.” The vast majority of cell types were much closer between humans and marmosets than between marmosets and mice. “That was very satisfying to see,” Krienen says. The cross-species study profiled the well-studied type, called Betz cells in humans. The team found an analogous cell in mice, reflecting common evolutionary origins, but electrical and some other properties differed markedly between species. “The mouse has some general similarities to a human, in terms of its body plan, but the details are different. The same is true at the level of cell types,” Lein says. “You have all the same types, with a few exceptions, but their properties change a bit, that’s the nature of our species differences.” By contrast, “chandelier” cells, named for their beautifully elaborate connection structures, are very similar across species.
The data will allow researchers to target specific cell types, using either long-established genetic engineering “transgenic” tools in mice, or, in other animals, DNA sequences delivered by harmless viruses. “The transgenic approach is effective for the well-established generation of mouse models,” says Krienen. “Viral-based tools, which can of course also be used in mice, really reach their potential as ways of delivering genes, regulatory elements or mutations in animals, for which we lack that genetic toolbox, like nonhuman primates.” Being able to target cell types like this will enable a wealth of new tools for everything from studying brain development to dissecting neural circuits. “Now we know which genes might be deployed differently from one cell type to another, we can build tools with the cell-type precision we’ve longed to,” Krienen says.
Understanding which genes and genetic sequences that regulate their activity are specific to different cell types will also advance researchers’ understanding of disease. “This is going to have a big impact on disease, because now we can pinpoint it to anatomy,” Lein says. “Where are the cells being impacted by a genetic mutation?” Knowing how similar disease-relevant features are in different species could also inform choices about animal models. That’s a major question that overhangs biological research; for example, is a study in mice relevant to humans? “If the relevant regulatory elements aren’t conserved, is a mouse model of schizophrenia ever going to yield the insights we hope to get?” says Krienen.
The varied reports represent a bumper crop of data, but important details are lacking. “What’s really missing here, that will be crucial, is proteins,” says neuroscientist Botond Roska, of the University of Basel, who was not involved in the project (but who advises the Allen Institute). “The only reason we have genes is because they code for proteins, this is the final machinery of cells.” Proteomics technologies exist, but not yet at single-cell resolution. It is also not clear what influence different conditions might have on these data. “There’s a massive influence of activity on gene expression,” says Roska. “You’d have to probe these brains in different states to show these cell types remain the same under different conditions.” These contributions, he says, are just a beginning. “It’s a very important first step, but it’s a long road to really standardize cell types in the brain,” Roska says. “This is the first draft; it’s a reasonable hypothesis, but now it’s ready to be scrutinized by the whole community, questioned, tested and refined.”
In the immediate term, the project is working on embedding data in 3-D space. “An atlas isn’t just a bunch of GPS coordinates; it’s having them located on a map,” says Bhaduri. “That will be transformative, because where cells are located in the brain is really important, and there’s a lot we don’t understand about how space and function interact.” Looking to the future, the project’s next stage, a huge effort called BICAN (BRAIN Initiative Cell Atlas Network), that aspires to move into nonhuman primates and humans, is already funded. “We’ve been able to really tackle the complexity of this one part of the brain,” Lein says. “Now the stage is set to extend this, both across the rest of the mouse brain, but also moving to nonhuman primates and the whole human brain.”


